Metadata isn’t just for the data

Honestly I didn’t learn this today, it’s something I’ve always had in the back of my head. But I just wanted to emphasize on how important it is to document everything around your research, not just the raw data, but the context, the commands, the decisions. That’s metadata too.
I’m a Bioinformatics PhD student, and I just wanted to share my humble experience on what I do to document my work.
What do I document?
Honestly? Everything.
Not just code, but all the tricky setup steps that no one talks about. Things like how to configure your laptop for bioinformatics work, set up the office printer (which could be tricky sometimes), or access the HPC cluster.
These are the kinds of things I was lucky to get help with early on, my supervisor guided me through a lot of the setup. But timing doesn’t always work out like that for everyone, so I started documenting these steps to make things easier for the next person (and for future me too).
It’s true that I sometimes forget to document in the moment. But every time I revisit my notes and they save me, I’m reminded just how valuable good documentation is.
So, no, it doesn’t take much time to just document on the fly. And trust me: “later” sometimes might never happen.
How do I document?
☀️ Start with clarity
I start every project with a context block, where I describe the aim of the analysis and the experimental design. It helps me actually understand what we’re trying to find or observe. This one might be personal since I’m also a biologist by training, so I like to make sure there’s a clear link between the data and the biological insight.
I always write documentation as if I know nothing about what I’m documenting. That forces me to include as many details as possible, so when future me checks the notes, it’s guaranteed to be understandable.
I also document failed experiments, and what I did to save the day. This is especially helpful in case someone else runs into the same issues, and honestly, even for me when I run into similar problems later.
📁 Stay organized
- I have a consistent folder structure (I personally use Google Drive and SeaFile, but I’ve seen people use other clouds, save locally, on a server, and that’s all fine). This helps me document on the fly without overthinking where to save or how to name things.
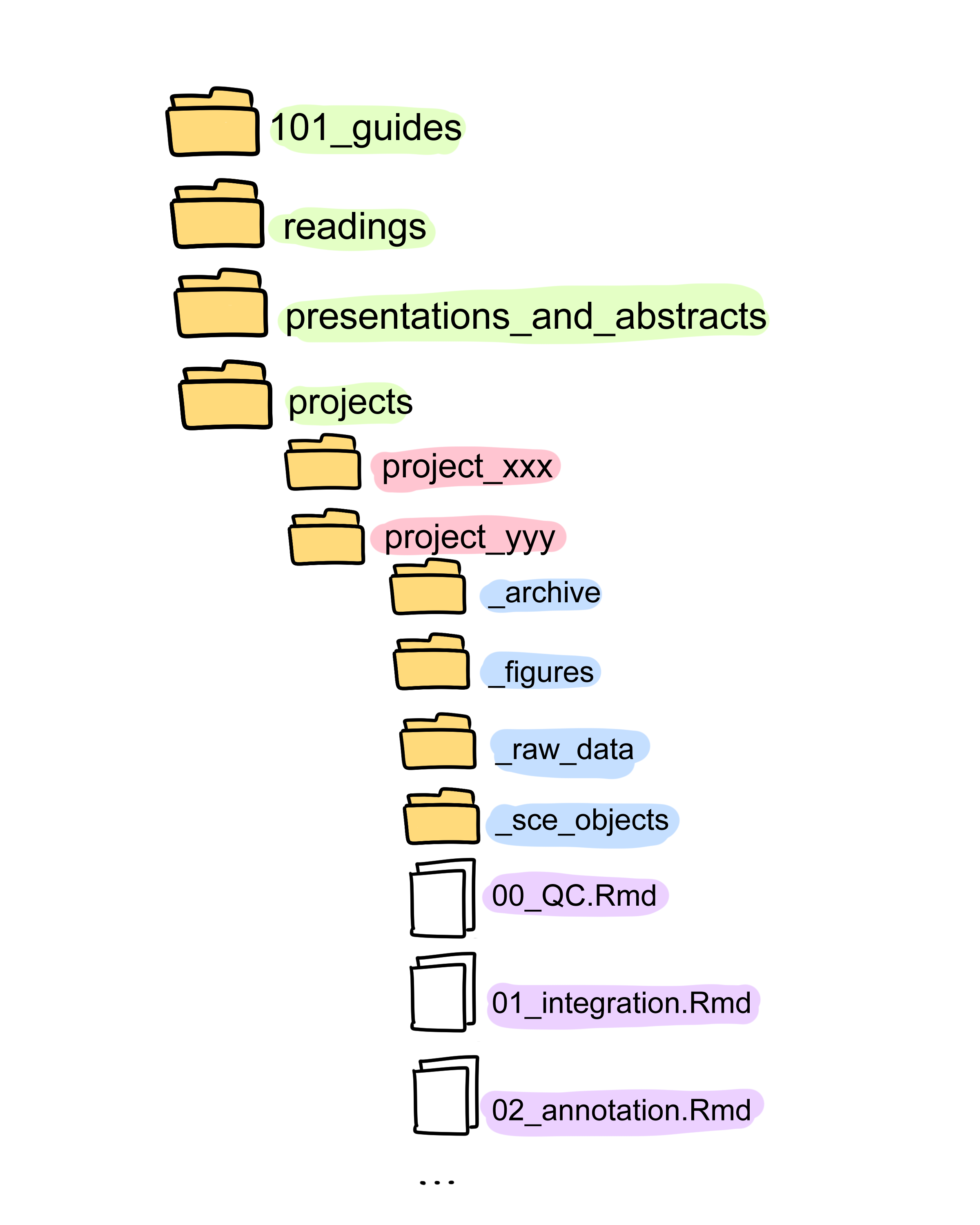
- Most of my learning notes include “101” in the file name. So when I need to find something quickly, I just type 101 in the search bar and rely on autocompletion.
→ So yeah, give your notes meaningful & searchable names!
I number my R Markdown files so that, in the future (or for collaborators), it’s clear which one to check first. I usually separate them by pipeline steps, e.g one .
Rmdfor preprocessing scRNAseq data, one for integration, one for annotation and so on. So if I have files like04-1-DE_analysis.Rmdand04-2-Annotate_Monocytes.Rmd, I know the annotation came after the first round of Differential Expression Analysis.A cool idea I learned from my supervisor: create a README file for each project. Include all the relevant info, e.g. where the raw data is stored, how to run the scripts/notebooks, and what decisions were made.
In the end, it’s not the longest list of tips, but I hope it’s enough to spark some inspiration and help you find what works best for you 😊
This is my first blog entry using Quarto, and nothing is perfect the first time XD. What matters is taking action 🤗
Stay safe, and keep documenting ❤️